Research interests:
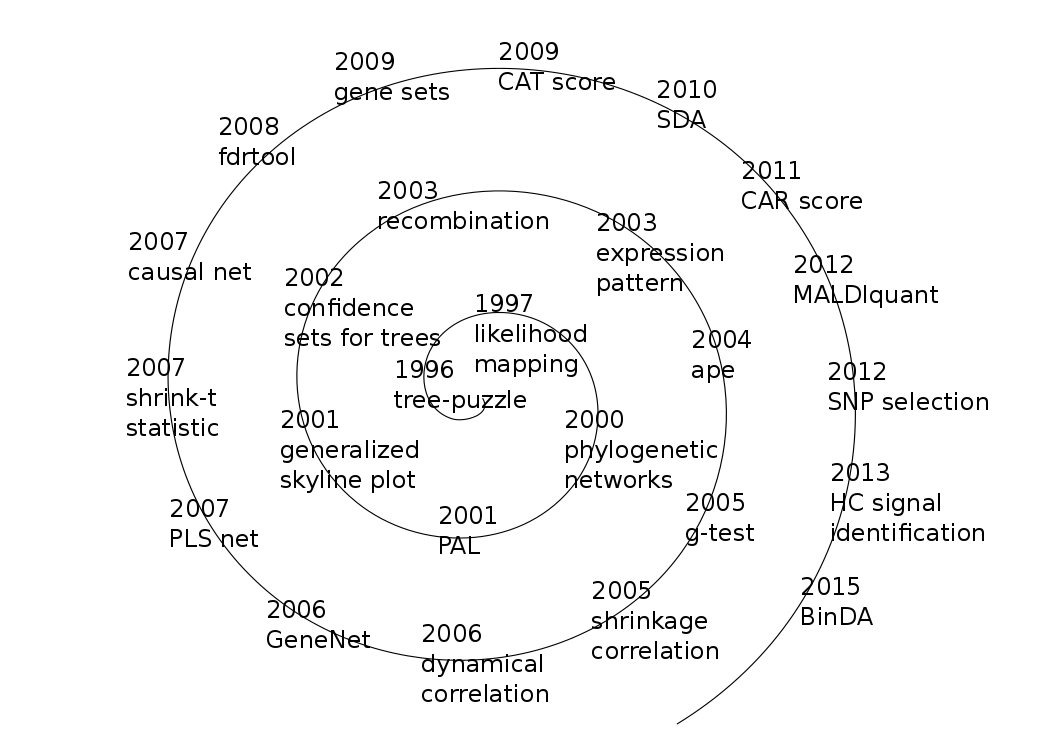
Timeline of methods developed in our group (click image for a larger view)
Our research focuses on statistical and machine learning methods and models for high-dimensional data and to understand the underlying principles of learning from data.
We have developed computationally effective methods for high-dimensional regularized estimation using shrinkage methods, for model and variable selection, for signal identification, for computing false discovery rates and performing dimension reduction, for learning graphical models and for regularized classification and regression.
Biological applications of our methods have included the analysis of transcriptomics and proteomics data, gene ranking and biomarker discovery, construction of biological networks and trees as well as modeling of molecular evolution.
Korbinian Strimmer is recognized as "Highly Cited Researcher".Our aim is to make all our work accessible and reproducible by providing corresponding free and open source software as well as analysis scripts and data sets.
R packages - statistical methods:
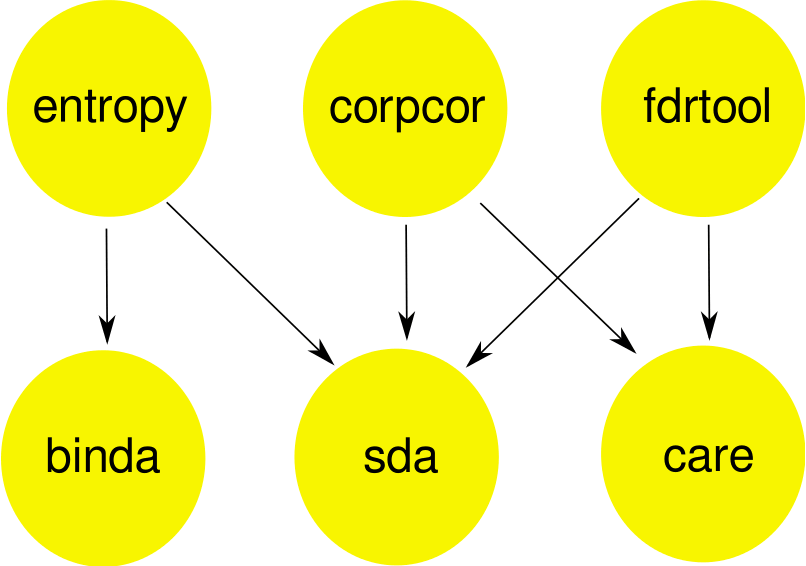
Statistical toolbox for analyzing high-dimensional data (click image for a larger view)
- binda: Multi-class discriminant analysis using binary predictors.
- care: High-dimensional regression and CAR score variable selection.
- corpcor: Efficient estimation of covariance and (partial) correlation.
- crossval: Generic functions for cross validation.
- entropy: Estimation of entropy, mutual information and other related quantities.
- fdrtool: Estimation of (local) false discovery rates and higher criticism.
- longitudinal: Analysis of multiple time course data.
- sda: Shrinkage discriminant analysis and CAT score variable selection.
- st: Shrinkage t statistic and CAT score.
- whitening: Whitening and high-dimensional canonical correlation analysis.
R packages - computational biology:
- GeneCycle: Identification of periodically expressed genes.
- GeneNet: Modeling and inferring gene networks.
- MALDIquant: Quantitative analysis of mass spectrometry data.
- MALDIquantForeign: Import/export routines for MALDIquant.
- readBrukerFlexData: Read MS data in Bruker *flex format.
- readMzXmlData: Read MS data in mzXML format.
- Contributions to ape (phylogenetics in R) and plsgenomics.
- R code for shrinkage estimation of VAR gene network.
Matlab/Octave code:
This code was kindly contributed by Kevin P. Murphy.- Shrinkage covariance and correlation estimation
(README):
covshrinkKPM.m (Matlab function),
testscript-matlab.txt, testscript-R.txt (test scripts),
smalldata.txt, largedata.txt (example data files).
Phylogenetics:
- TREE-PUZZLE: Maximum-likelihood tree reconstruction package.
- PAL: Java library for methods in molecular sequence analysis.
- Vanilla: Java command line programs based on PAL.
Most of the data provided below are packaged as part of an R package. For further information concerning the raw data please consult the original data authors and papers.
From Jendoubi and Strimmer, 2019 (CCA data integration):
- See the whitening R package. The Cancer Genome Atlas LUSC data: see lusc data.
- Martin et al. (2007) nutrimouse study: see nutrimouse data.
From Gibb and Strimmer, 2015 (differential proteomics):
- See the binda and MALDIquant R packages. Dorothea NIPS 2003 drug discovery data set: dorothea.rda.
- Fiedler et al. (2009) pancreas cancer proteomics data: spectra.tar.gz and spectra_info.csv
From Zuber and Strimmer, 2011 (CAR score):
- See the care R package. Lu et al. (2004) brain aging gene expression data: see lu2004 data.
From Zuber and Strimmer, 2009 (CAT score):
- See the sda R package. Sreekumar et al. (2009) Prostate cancer metabolomic data: sreekumar2009.rda.gz.
From Opgen-Rhein and Strimmer, 2007a (VAR network) and 2007b (approximate causal network):
- See the GeneNet R package. Smith et al. (2004) A. thaliana time series data: arth-smith2004.rda.gz.
- See also the R script for computing an approximate causal network and the R code for estimating the VAR network.
From Opgen-Rhein and Strimmer, 2007 (shrinkage t statistic):
- Cope et al. (2004) Affymetrix spike-in data: affyspike.rda.gz.
- See the st R package. Choe et al. (2005) "golden spike" experiment: see choedata.
- van 't Wout et al. (2003) HIV-1 data: vantwout-hiv1.rda.gz.
From Boulesteix and Strimmer, 2005 (PLS transcription factor prediction):
- Kao et al. (2004) Ecoli experiment: kaoecoli.rda.gz.
- Liao et al. (2003) hemoglobin data: hemoglobin.rda.gz.
- Spellmann et al. (1998) and Gasch et al. (2000) yeast data, with compatible Lee et al. (2002) ChIP matrix: yeast-tfa.rda.gz.
- Use this R script to check that you retrieved the complete set of data.
From Wichert, Fokianos and Strimmer, 2004 (identification of cell cycle genes):
- Cho et al. (2001) human fibrolast data: fibroblasts.rda.gz.
- See the GeneCycle R package. Laub et al. (2000) caulobacter data: caulobacter.
- Spellmann et al. (1998) yeast data: spellmann-yeast.rda.gz.
- Whitfield et al. (2002) human HeLa data: humanhela.rda.gz.
- Use this R script to check that you retrieved the complete set of data.
- Table 2 of Wichert et al. (2004) can be reproduced by using this R script.
From Strimmer, 2003 (quasi-likelihood approach):
- Golub et al. (1999) leukemia data: golub.rda.gz.
From diverse other papers:
- See the longitudinal R package. Rangel et al. (2004) human T-cell data: tcell.
- Hedenfalk et al. (2001) breast cancer data: hedenfalk.rda.gz.
- Hedges (1994) alignment: amniotes-seqs.txt and amniotes-info.txt.
- See the GeneNet R package. Schmidt-Heck et al. (2004) E. coli data: ecoli.
- Tanaka et al. (2004) HCV tree: hcv.tre (Newick tree).
- West et al. (2001) breast cancer data: westdataclean.rda.gz.
- See the ape R package. Yusim et al. (2001) HIV-1 tree: hivtree.
Our group was organizer of the meeting Statistical Methods for Postgenomic Data (SMPGD 2017) at Imperial College London. Previously, we helped organizing the life science session at GOCPS 2010 and the Computational Systems Biology (WCSB 2008) conference at the University of Leipzig. At LMU Munich our group was coorganizer of the workshop Complex Stochastic Systems in Biology and Medicine 2004.
